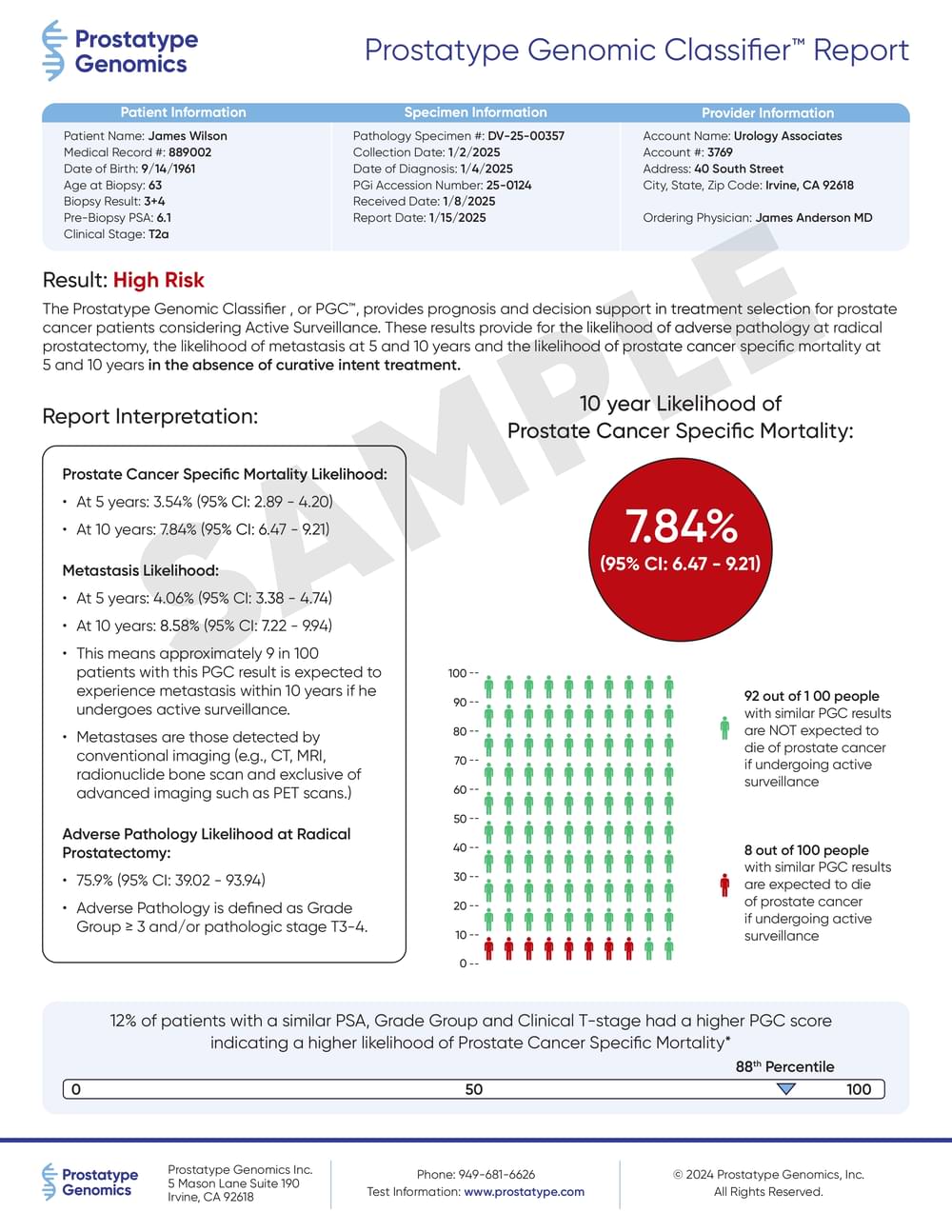
What is your patient’s Prostatype?
The Prostatype® Genomic ClassifierTM or PGCTM helps predict important clinical outcomes, in the absence of therapy, that can be used to guide Active Surveillance decisions. The test provides a personalized assessment of a patient’s risk for:
- 5- and 10-Year PCa Specific Mortality
- 5- and 10-Year Metastasis
- Adverse Pathology at Radical Prostatectomy
What is your patient’s Prostatype?
The Prostatype® genomic test helps guide Active Surveillance decisions. The test provides a personalized assessment of a patient’s risk for:
- 10-Year PCa Specific Mortality
- 10-Year Metastasis
- Adverse Pathology
Prostatype Genomics’ patented genomic test accurately determines the aggressiveness of prostate cancer with unparalleled precision.
The Prostatype® test is an indispensable tool for patients and their doctors in selecting the most appropriate treatment for each individual patient. The test enables highly personalized care, making it an essential component of treatment decision making.
Prostatype is a genomic test that provides a comprehensive assessment of a patient’s prostate cancer aggressiveness to help answer the question, Active Surveillance or Radical Therapy?
Using prostate tissue from the biopsy obtained at the time of diagnosis (no new sampling of the prostate is needed) the gene expression of the tumor is analyzed. This gene signature, combined with clinical information calculates your patient’s personalized result, providing you and your patient the information you need to make the right choice in care. With the help of the Prostatype Genomic Classifier you and your patient can make a better treatment decision and thus minimize the risk of over or under-treatment of the cancer.
The Prostatype Genomic Classifier Supports Your Treatment Decisions
- Improve patient risk stratification and treatment recommendations
- Increase peace of mind for men choosing active surveillance
- Reduce the risk of over and under treatment
Prostatype Genomic Classifier to Inform Patient Management
- Prostatype is a tissue-based molecular assay that measures the expression of 3 prostate cancer embryonic stem cell gene predictors.
- The Prostatype gene signature is combined with clinical parameters to generate an individualized risk assessment.
- The Prostatype Genomic Classifier has been extensively validated from biopsy tissue to help guide active surveillance decisions.
- The Prostatype Genomic Classifier is indicated for patients presently on, or considering, active surveillance.
- The Prostatype Genomic Classifier provides actionable information about a patient’s risk of adverse pathology at radical prostatectomy, 5- and 10-year risk of metastases and 5- and 10-year PCa-specific mortality in the absence of treatment.
1) A Novel Risk Score (P-Score) Based on a Three-Gene Signature, for Estimating the Risk of Prostate Cancer-Specific Mortality. Söderdahl et al, Res Rep Urol. 2022, 2) An expression signature at diagnosis to estimate prostate cancer patients’ overall survival. Peng et al, Prostate Cancer Prostatic Dis. 2014
The Prostatype Genomic Classifier Delivers Actionable Results to Guide Treatment Decisions
The Prostatype® Genomic Classifier provides prognostic information to help determine your patient’s eligibility for active surveillance.
Prostatype provides prognostic insights into a patient’s risk for adverse pathology, 5- and 10 year PCSM and 5- and 10-year metastasis for patients while on active surveillance to help you make more informed management decisions
Source: Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. Sæmundsson et al, The Prostate 2023
The Prostatype Genomic Classifier Helps Ensure Your Patients Get the Right Treatment at The Right Time
Prostatype has been clinically validated on biopsy tissue to improve patient risk classification.
In a recent study, Prostatype accurately re-classified patients initially classified using D’Amico criteria into low, intermediate and high-risk risk groups based on their 10-year prostate cancer-specific mortality risk (p-value <0.0001).
Source: Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. Sæmundsson et al, The Prostate 2023

The Prostatype Genomic Classifier Helps You Confidently Choose Active Surveillance
- 0.93 AUC for 10 Yr PCa Mortality
- 0.88 AUC for 10 Yr Metastatic Risk
- 0.81 AUC for Adverse Pathology*
*Precise risk stratification of prostate cancer by P-score in an Asian population. See-Tong Pang et al
Prostatype Helps You Confidently Choose Active Surveillance
- 0.93 AUC for 10 Yr PCa Mortality
- 0.88 AUC for 10 Yr Metastatic Risk
- 0.81 AUC for Adverse PathologyZ*
*Precise risk stratification of prostate cancer by P-score in an Asian population. See-Tong Pang et al
Prostatype® Outperforms Nomograms on Prediction of 10-year Prostate Cancer-Specific Mortality

Prostatype® Outperforms Nomograms on Prediction of 10-year Prostate Cancer-Specific Mortality
In a study of 316 patients analyzed with median follow-up of 8.8 yrs, the Prostatype P-Score improved the prediction of prostate cancer-specific death compared to CAPRA (p=0.05) and D’Amico (p=0.001):
- P-Score (AUC=0.93, 95% CI:0.89-0.98)
- CAPRA (AUC=0.88, 95% CI:0.80-0.96)
- D’Amico (AUC=0.81, 95% CI:0.72-0.90)
Source: Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. Sæmundsson et al, The Prostate 2023

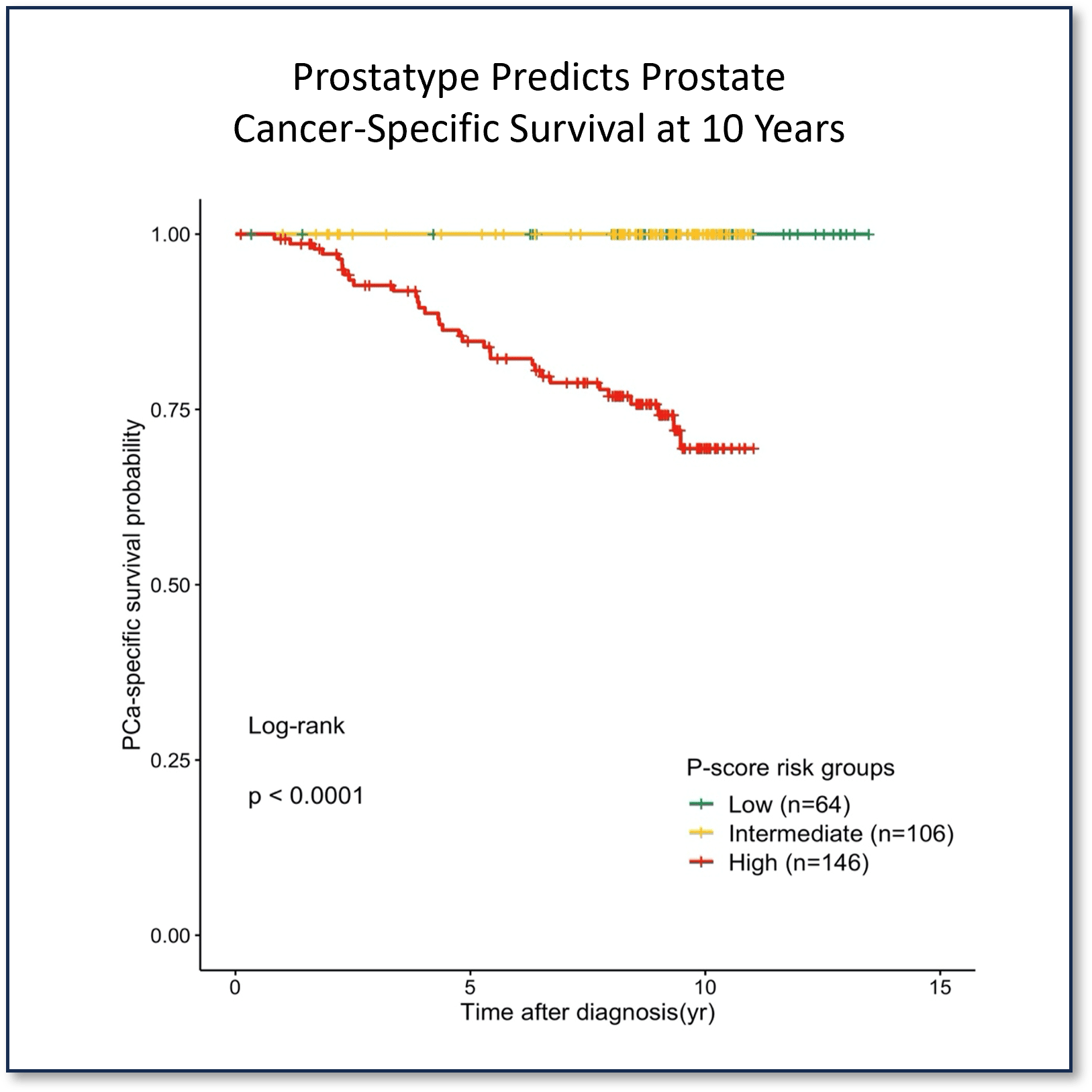
Prostatype® Helps Identify the Right Candidates for Active Surveillance
Prostatype accurately stratified all 316 patients into Low-, Intermediate- and High-risk groups for prostate cancer-specific survival at a significant p-value <0.0001.
None of the patients in the Prostatype low or intermediate risk groups group died from prostate cancer during follow-up.
Source: Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. Sæmundsson et al, The Prostate 2023
Prostatype® Provides Insights into Your Patient’s Metastatic Risk
Prostatype accurately categorized all 316 patients into Low-, Intermediate- and High-risk groups for metastasis-free survival at a statistically significant p-value <0.0001.
Source: Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. Sæmundsson et al, The Prostate 2023

Prostatype® Stratifies Patients at Risk for Harboring Adverse Pathology
Prostatype helps improve the assessment of patients considering active surveillance. Utilizing tissue from the core needle biopsy, Prostatype predicted:
- pT-stage ≥T3a (odds ratio=1.3, 95% CI: 1.2-1.4, p<0.0001)
- ISUP grades ≥3 (odds ratio=1.5, 95% CI: 1.3-1.9, p<0.0001) discovered in RP specimens
The median Prostatype values were significantly different in pT-stage groups (Chi-Square=22.2, p<0.0001) as well as in ISUP grades (Chi-Square=29.8, p<0.0001) USING Kruskal-Wallis analysis.
Source: Validation of the prognostic value of a three-gene signature and clinical parameters-based risk score in prostate cancer patients. Sæmundsson et al, The Prostate 2023

Prostatype® Genes and Technology Reported in Multiple Studies on Over 1,880 Patients
2026
2023
2023
2022
2016
2014
2014
Getting Started with The Prostatype Genomic Classifier
For more information about the Prostatype Genomic Classifier please contact us and our customer care team:
To view sample patient reports, please follow the link below:
To set up an account with Prostatype Genomics, please complete the following setup form provided within this link:
To order sample collection kits, please follow the link below:
To download and print a copy of the test requisition form, please follow the link below:
To obtain a copy of the specimen requirements and test submission instructions, please follow the link below:
Before Deciding Know His Prostatype!